
DNA replication (slides)
DNA strands separate and then these strands are used as a template ; idea suggested by Watson and Crick called the semi-conservative model
The semi-conservative model was proven by Meselson and Stahl
Enzymes used in DNA replication
Gyrase - relieves tension of the unwinding of the double helix
Helicase -break H bonds of complementary strands
Primase - Make RNA primer ( ~10 nucleotides long in eukaryotes )
DNA Polymerases - Add DNA nucleotides to new strand ( I, II,II, in prokaryotes 5 diff types in eukaryotes)
Ligases - attach Okazaki fragments on the lagging strand
Rate of nucleotides added
The rate of nucleotides added at a rate of 50/s in eukaryotes
The rate of nucleotides added at a rate of 500/s in prokaryotes
The source of Energy for DNA replication is Nucleoside triphosphate
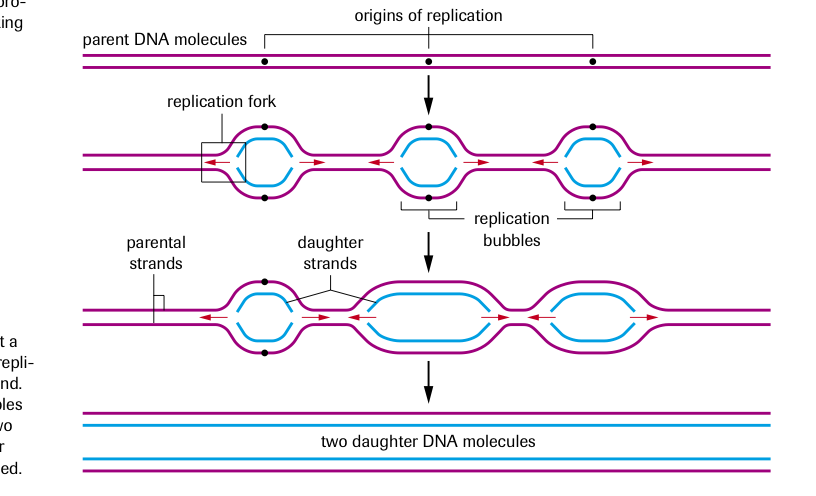
In eukaryotes DNA replication occurs at more than one site at a time, resulting in hundreds of replication forks across a DNA strand.
Eventually the replication bubbles become continuous and the two new double stranded daughter molecules are completely formed
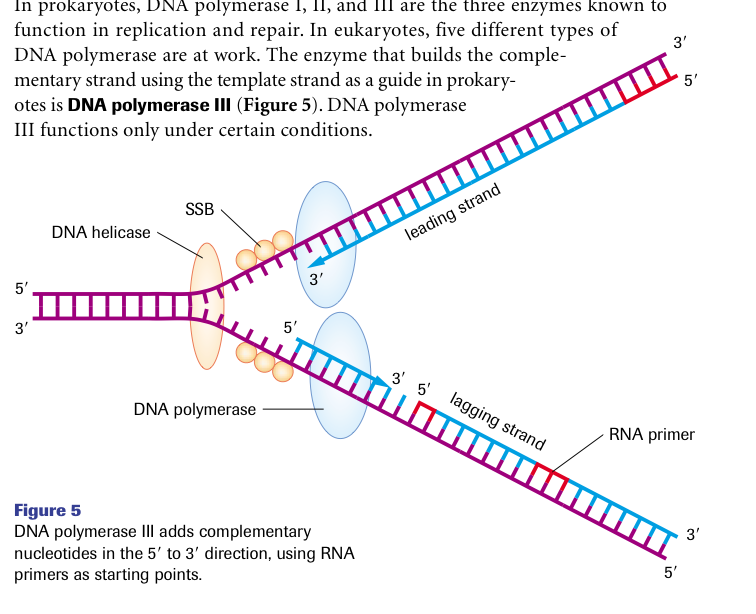
DNA polymerase III adds complementary nucleotides in the 5’ to 3’ direction using RNA primers as starting points
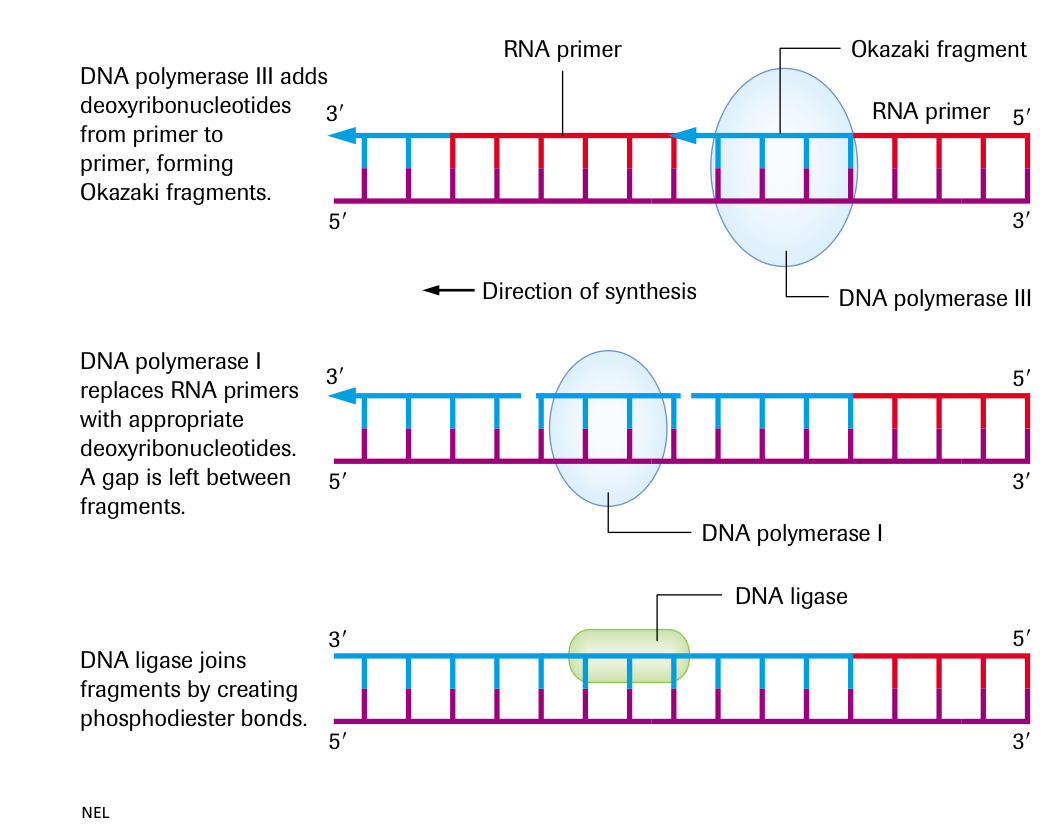
DNA polymerase II adds deoxyribonucleotides from primer to primer, forming Okazaki fragments
DNA polymerase I replaces RNA primers with appropriate deoxyribonucleotides. A gap is left between fragments
DNA ligase joins fragments by creating phosphodiester bonds
DNA is replicated 5’ to 3’
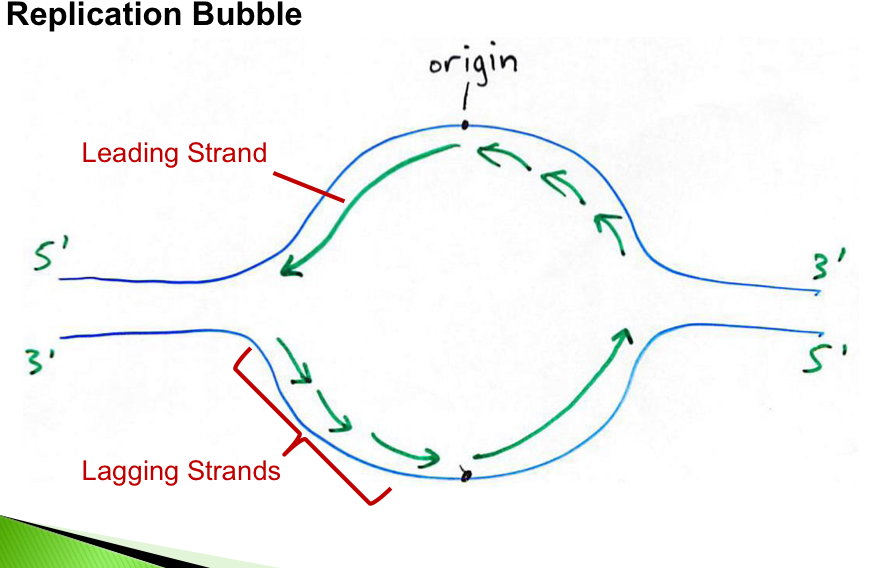
replication bubble
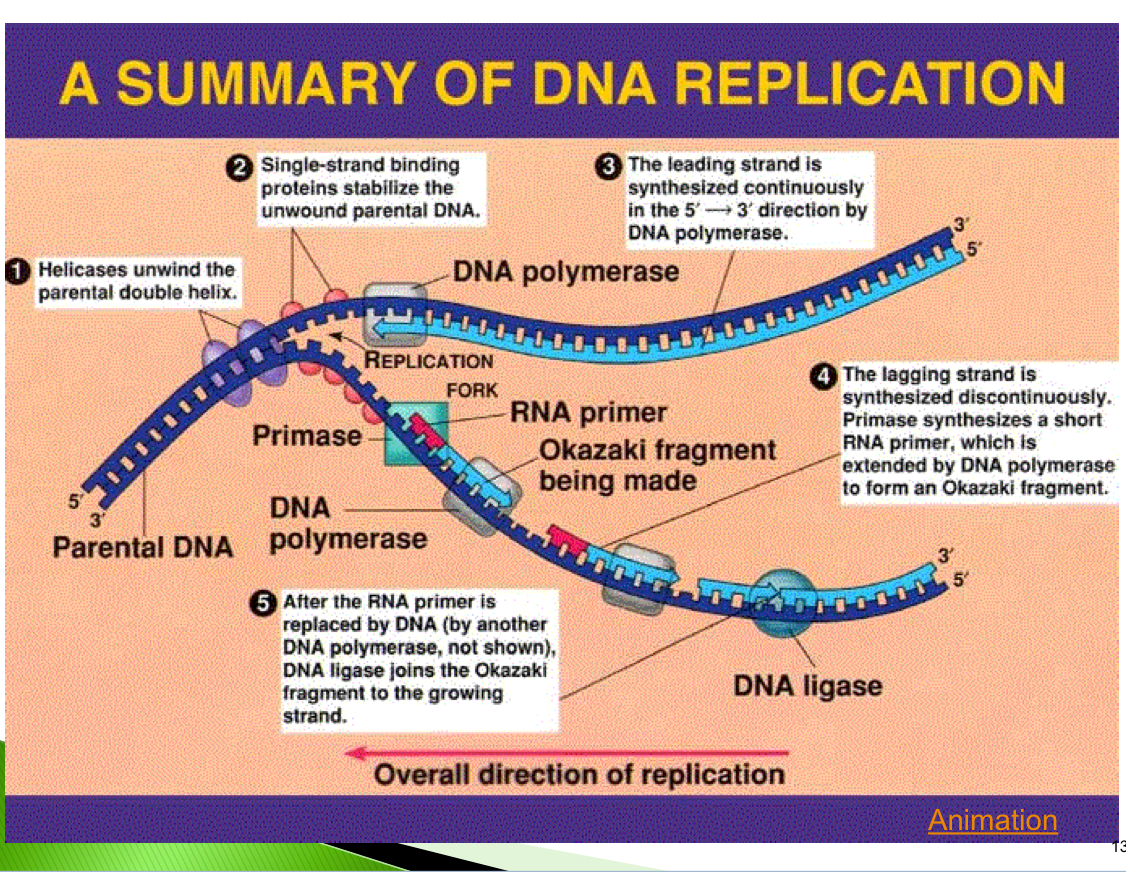
A summary of DNA replication
Helicase unwind the parental double helix
single strand binding proteins stabilize the unwound parental DNA
the leading strand is synthesized continuously
the lagging strand is synthesized discontinuously Primase synthesizes a short RNA primer, which is extend by DNA polymerase to form an Okazaki fragment
After the RNA primer is replaced by DNA ( by another DNA polymerase, not shown) DNA ligase joins the Okazaki fragments to the growing strand
Synthesis of Leading strand | Synthesis of lagging Strand |
|---|---|
Priming (primase) | Priming for Okazaki Fragment ( primase) |
Elongation (DNA polymerase III) | Elongation of fragment (DNA polymerase III) |
Replacement of RNA primer by DNA (DNA polymerase I) | Replacement of RNA primer by DNA (DNA polymerase I) |
Replication towards the fork | Replication away from the fork |
DNA proofreading
DNA polymerase proofreads (I and III)
check each nucleotide against the template as it is added
if there is a mistake, it backs up and removes incorrect nucleotide and repplaces it before continuing ( exonuclease)
DNA repair
DNA replication (slides)
DNA strands separate and then these strands are used as a template ; idea suggested by Watson and Crick called the semi-conservative model
The semi-conservative model was proven by Meselson and Stahl
Enzymes used in DNA replication
Gyrase - relieves tension of the unwinding of the double helix
Helicase -break H bonds of complementary strands
Primase - Make RNA primer ( ~10 nucleotides long in eukaryotes )
DNA Polymerases - Add DNA nucleotides to new strand ( I, II,II, in prokaryotes 5 diff types in eukaryotes)
Ligases - attach Okazaki fragments on the lagging strand
Rate of nucleotides added
The rate of nucleotides added at a rate of 50/s in eukaryotes
The rate of nucleotides added at a rate of 500/s in prokaryotes
The source of Energy for DNA replication is Nucleoside triphosphate

In eukaryotes DNA replication occurs at more than one site at a time, resulting in hundreds of replication forks across a DNA strand.
Eventually the replication bubbles become continuous and the two new double stranded daughter molecules are completely formed

DNA polymerase III adds complementary nucleotides in the 5’ to 3’ direction using RNA primers as starting points

DNA polymerase II adds deoxyribonucleotides from primer to primer, forming Okazaki fragments
DNA polymerase I replaces RNA primers with appropriate deoxyribonucleotides. A gap is left between fragments
DNA ligase joins fragments by creating phosphodiester bonds
DNA is replicated 5’ to 3’
replication bubble

A summary of DNA replication
Helicase unwind the parental double helix
single strand binding proteins stabilize the unwound parental DNA
the leading strand is synthesized continuously
the lagging strand is synthesized discontinuously Primase synthesizes a short RNA primer, which is extend by DNA polymerase to form an Okazaki fragment
After the RNA primer is replaced by DNA ( by another DNA polymerase, not shown) DNA ligase joins the Okazaki fragments to the growing strand

Synthesis of Leading strand | Synthesis of lagging Strand |
|---|---|
Priming (primase) | Priming for Okazaki Fragment ( primase) |
Elongation (DNA polymerase III) | Elongation of fragment (DNA polymerase III) |
Replacement of RNA primer by DNA (DNA polymerase I) | Replacement of RNA primer by DNA (DNA polymerase I) |
Replication towards the fork | Replication away from the fork |
DNA proofreading
DNA polymerase proofreads (I and III)
check each nucleotide against the template as it is added
if there is a mistake, it backs up and removes incorrect nucleotide and repplaces it before continuing ( exonuclease)
DNA repair
 Knowt
Knowt